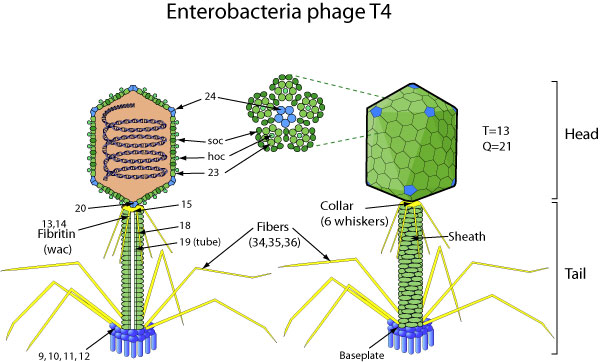
Introduction
Phage T4 has provided innumerable contributions to the paradigms of genetics and biochemistry. Its complete genome sequence of 168,903 bp encodes around 300 gene products. The biology of T4 and its genomic sequence provide the best-understood model for modern functional genomics and proteomics. Variations in gene expression, including overlapping genes, internal translation initiation, spliced genes, translation drift, and RNA processing, alert us to the caveats of purely computational methods.
The transcriptional pattern of T4 reflects its dependence on host RNA polymerase and the use of phage-encoded proteins that sequentially modify the RNA polymerase; Transcriptional activator proteins, a phage sigma factor, anti-sigma, and sigma decoy proteins also act to specify early, mid, and late promoter recognition. Post-transcriptional controls of Enterobacteria phage T4 Recombinants provide excellent systems for the study of RNA-dependent processes, particularly at the structural level.
The redundancy of T4’s DNA replication and recombination systems reveals how phage and other genomes stably replicate and repair in different environments, providing insights into genome evolution and adaptations to new hosts and growth environments. . In addition, genomic sequence analysis has provided new insights into tail fibre variation, lysis, gene duplications, and membrane protein localization, while the high-resolution structure determination of the “device of cell perforation”, combined with three-dimensional image reconstruction of the baseplate, has revealed the mechanism of penetration during infection.
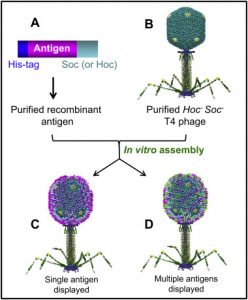
Despite these advances, nearly 130 potential T4 genes remain uncharacterized. Current phage sequencing initiatives are now revealing the similarities and differences between members of the T4 family, including those that infect bacteria other than Escherichia coli. T4 functional genomics will aid in the interpretation of these recently sequenced T4-related genomes and in expanding our understanding of the complex evolution and ecology of phages, the most abundant and among the oldest biological entities on Earth.
Quality level: 100
Biological source: Escherichia coli (NM 989)
Description: a recombinant form of the phage T4 enzyme
Form: solution
Specific activity: ~3000 units/mg protein
Packaging
- 100U pack (10481220001 [1U/μl])
- 500U pack (10716359001 [1U/μl])
- 500U pack (10799009001 [5U/μl])
Manufacturer/trade name: Roche
Parameter: 4-22 °C optimum reaction temperature.
Technique(s)
Activity test: adequate
Optimum pH: 7.2-7.8
General description
T4 DNA ligase catalyzes the formation of phosphodiester bonds between neighbouring 3′-hydroxyl and 5′-phosphate ends in double-stranded DNA. Single-stranded nicks in double-stranded DNA are also closed by T4 DNA ligase.
Content
T4 DNA Ligase, supplied with 10x concentrated ligation buffer including ATP.
Specificity
Heat inactivation: The enzyme can be inactivated by heating (65°C, 10 minutes).
Request
Ligation of DNA fragments with sticky and blunt ends.
Quality
Tested for the absence of deoxyribonucleases and exonucleases, in accordance with current Quality Control procedures.
Unit Definition
One unit of T4 DNA ligase is the amount of enzymatic activity that converts 1 nmol of 32P pyrophosphate to Norit-absorbable material in 20 minutes at +37°C. One unit corresponds to 0.2 U determined in the exonuclease III resistance assay. Dilution buffer: 50 mM Tris-HCl, 10 mM dithioerythritol, bovine serum albumin, 500 μg/ml, pH 7.6 (+25 °C).
Volume Activity: 1 x 103 U/ml; 5x103U/mL
Preparation note
- Activator: Mg2+ (10mM), DTT
- Working solution: For enzyme dilution, Roche recommends using a buffer containing the storage buffer components: 20 mM Tris-HCl, 60 mM KCl, 1 mM EDTA, 5 mM ditoerythritol, 50% glycerol (v/ v), pH 7.5 (4°C).
Other notes
For life science research only. It should not be used in diagnostic procedures.